Instance Segmentation Module Tutorial¶
I. Overview¶
The instance segmentation module is a crucial component in computer vision systems, responsible for identifying and marking pixels that contain specific object instances in images or videos. The performance of this module directly impacts the accuracy and efficiency of the entire computer vision system. The instance segmentation module typically outputs pixel-level masks (masks) for each target instance, which are then passed as input to the object recognition module for subsequent processing.
II. Supported Model List¶
The inference time only includes the model inference time and does not include the time for pre- or post-processing.
| Model | Model Download Link | Mask AP | GPU Inference Time (ms) [Normal Mode / High-Performance Mode] |
CPU Inference Time (ms) [Normal Mode / High-Performance Mode] |
Model Storage Size (MB) | Description |
|---|---|---|---|---|---|---|
| Mask-RT-DETR-H | Inference Model/Training Model | 50.6 | 180.83 / 180.83 | 1711.24 / 1711.24 | 449.9 | Mask-RT-DETR is an instance segmentation model based on RT-DETR. By adopting the high-performance PP-HGNetV2 as the backbone network and constructing a MaskHybridEncoder encoder, along with introducing IOU-aware Query Selection technology, it achieves state-of-the-art (SOTA) instance segmentation accuracy with the same inference time. |
| Mask-RT-DETR-L | Inference Model/Training Model | 45.7 | 113.20 / 113.20 | 1179.56 / 1179.56 | 113.6 |
❗ The above list features the 2 core models that the image classification module primarily supports. In total, this module supports 15 models. The complete list of models is as follows:
👉Model List Details
| Model | Model Download Link | Mask AP | GPU Inference Time (ms) [Normal Mode / High-Performance Mode] |
CPU Inference Time (ms) [Normal Mode / High-Performance Mode] |
Model Storage Size (MB) | Description |
|---|---|---|---|---|---|---|
| Cascade-MaskRCNN-ResNet50-FPN | Inference Model/Training Model | 36.3 | 136.79 / 136.79 | - / 5935.41 | 254.8 | Cascade-MaskRCNN is an improved Mask RCNN instance segmentation model that utilizes multiple detectors in a cascade, optimizing segmentation results by leveraging different IOU thresholds to address the mismatch between detection and inference stages, thereby enhancing instance segmentation accuracy. |
| Cascade-MaskRCNN-ResNet50-vd-SSLDv2-FPN | Inference Model/Training Model | 39.1 | 137.40 / 137.40 | - / 6816.68 | 254.7 | |
| Mask-RT-DETR-H | Inference Model/Training Model | 50.6 | 180.83 / 180.83 | 1711.24 / 1711.24 | 449.9 | Mask-RT-DETR is an instance segmentation model based on RT-DETR. By adopting the high-performance PP-HGNetV2 as the backbone network and constructing a MaskHybridEncoder encoder, along with introducing IOU-aware Query Selection technology, it achieves state-of-the-art (SOTA) instance segmentation accuracy with the same inference time. |
| Mask-RT-DETR-L | Inference Model/Training Model | 45.7 | 113.20 / 113.20 | 1179.56 / 1179.56 | 113.6 | |
| Mask-RT-DETR-M | Inference Model/Training Model | 42.7 | 87.08 / 87.08 | - / 2090.73 | 66.6 | |
| Mask-RT-DETR-S | Inference Model/Training Model | 41.0 | 120.86 / 120.86 | - / 2163.07 | 51.8 | |
| Mask-RT-DETR-X | Inference Model/Training Model | 47.5 | 141.43 / 141.43 | 1379.14 / 1379.14 | 237.5 | |
| MaskRCNN-ResNet50-FPN | Inference Model/Training Model | 35.6 | 112.79 / 112.79 | - / 4912.37 | 157.5 | Mask R-CNN is a full-task deep learning model from Facebook AI Research (FAIR) that can perform object classification and localization in a single model, combined with image-level masks to complete segmentation tasks. |
| MaskRCNN-ResNet50-vd-FPN | Inference Model/Training Model | 36.4 | 112.88 / 112.88 | - / 5204.97 | 157.5 | |
| MaskRCNN-ResNet50 | Inference Model/Training Model | 32.8 | 181.60 / 181.60 | - / 5523.45 | 128.7 | |
| MaskRCNN-ResNet101-FPN | Inference Model/Training Model | 36.6 | 138.84 / 138.84 | - / 5107.74 | 225.4 | |
| MaskRCNN-ResNet101-vd-FPN | Inference Model/Training Model | 38.1 | 141.73 / 141.73 | - / 5592.76 | 225.1 | |
| MaskRCNN-ResNeXt101-vd-FPN | Inference Model/Training Model | 39.5 | 220.83 / 220.83 | - / 5932.59 | 370.0 | |
| PP-YOLOE_seg-S | Inference Model/Training Model | 32.5 | 243.41 / 222.30 | 2507.70 / 1282.35 | 31.5 | PP-YOLOE_seg is an instance segmentation model based on PP-YOLOE. This model inherits PP-YOLOE's backbone and head, significantly enhancing instance segmentation performance and inference speed through the design of a PP-YOLOE instance segmentation head. |
| SOLOv2 | Inference Model/Training Model | 35.5 | 131.99 / 131.99 | - / 2369.98 | 179.1 | SOLOv2 is a real-time instance segmentation algorithm that segments objects by location. This model is an improved version of SOLO, achieving a good balance between accuracy and speed through the introduction of mask learning and mask NMS. |
- Performance Test Environment
- Test Dataset:COCO2017 validation set.
- Hardware Configuration:
- GPU: NVIDIA Tesla T4
- CPU: Intel Xeon Gold 6271C @ 2.60GHz
- Software Environment:
- Ubuntu 20.04 / CUDA 11.8 / cuDNN 8.9 / TensorRT 8.6.1.6
- paddlepaddle 3.0.0 / paddlex 3.0.3
| Mode | GPU Configuration | CPU Configuration | Acceleration Technology Combination |
|---|---|---|---|
| Normal Mode | FP32 Precision / No TRT Acceleration | FP32 Precision / 8 Threads | PaddleInference |
| High-Performance Mode | Optimal combination of pre-selected precision types and acceleration strategies | FP32 Precision / 8 Threads | Pre-selected optimal backend (Paddle/OpenVINO/TRT, etc.) |
III. Quick Integration¶
❗ Before quick integration, please install the PaddleX wheel package. For detailed instructions, refer to the PaddleX Local Installation Tutorial
After installing the wheel package, a few lines of code can complete the inference of the instance segmentation module. You can switch models under this module freely, and you can also integrate the model inference of the instance segmentation module into your project. Before running the following code, please download the demo image to your local machine.
from paddlex import create_model
model = create_model("PP-YOLOE_seg-S")
output = model.predict("general_instance_segmentation_004.png", batch_size=1)
for res in output:
res.print()
res.save_to_img("./output/")
res.save_to_json("./output/res.json")
After running, the result obtained is:
{'res': "{'input_path': 'general_instance_segmentation_004.png', 'page_index': None, 'boxes': [{'cls_id': 0, 'label': 'person', 'score': 0.8723232746124268, 'coordinate': [88.34339, 109.87673, 401.85236, 575.59576]}, {'cls_id': 0, 'label': 'person', 'score': 0.8711188435554504, 'coordinate': [325.114, 1.1152496, 644.10266, 575.359]}, {'cls_id': 0, 'label': 'person', 'score': 0.842758297920227, 'coordinate': [514.18964, 21.760618, 768, 576]}, {'cls_id': 0, 'label': 'person', 'score': 0.8332827091217041, 'coordinate': [0.105075076, 0, 189.23515, 575.9612]}], 'masks': '...'}"}
The meanings of the running results parameters are as follows:
- input_path: Indicates the path of the input image to be predicted.
- page_index: If the input is a PDF file, it represents the current page number of the PDF; otherwise, it is None.
- boxes: Information of each detected object.
- cls_id: Class ID.
- label: Class name.
- score: Prediction score.
- coordinate: Coordinates of the bounding box, in the format [xmin, ymin, xmax, ymax].
- pred: The actual mask predicted by the instance segmentation model. Since the data is too large to be printed directly, it is replaced with ... here. You can use res.save_to_img() to save the prediction results as an image and res.save_to_json() to save the prediction results as a JSON file.
The visualization image is as follows:

Note: Due to network issues, the above URL may not be accessible. If you need to access this link, please check the validity of the URL and try again. If the problem persists, it may be related to the link itself or the network connection.
Related methods, parameters, and explanations are as follows:
create_modelinstantiates a general instance segmentation model (here,PP-YOLOE_seg-Sis used as an example), and the specific explanations are as follows:
| Parameter | Parameter Description | Parameter Type | Options | Default Value |
|---|---|---|---|---|
model_name |
Name of the model | str |
None | None |
model_dir |
Path to store the model | str |
None | None |
device |
The device used for model inference | str |
It supports specifying specific GPU card numbers, such as "gpu:0", other hardware card numbers, such as "npu:0", or CPU, such as "cpu". | gpu:0 |
threshold |
Threshold for filtering low-confidence objects | float/None |
None | None |
use_hpip |
Whether to enable the high-performance inference plugin | bool |
None | False |
hpi_config |
High-performance inference configuration | dict | None |
None | None |
- The
model_namemust be specified. After specifyingmodel_name, the default model parameters built into PaddleX are used. Ifmodel_diris specified, the user-defined model is used. -
thresholdis the threshold for filtering low-confidence objects. The default isNone, which means using the settings from the lower priority. The priority of parameter settings from highest to lowest is:predict parameter > create_model initialization > yaml configuration file. -
The
predict()method of the general instance segmentation model is called for inference prediction. Thepredict()method has parametersinput,batch_size, andthreshold, which are explained as follows:
| Parameter | Parameter Description | Parameter Type | Options | Default Value |
|---|---|---|---|---|
input |
Data to be predicted, supporting multiple input types | Python Var/str/list |
|
None |
batch_size |
Batch size | int |
Any integer | 1 |
threshold |
Threshold for filtering low-confidence objects | float/None |
|
None |
- The prediction results are processed, and the prediction result for each sample is of type
dict. It supports operations such as printing, saving as an image, and saving as ajsonfile:
| Method | Method Description | Parameter | Parameter Type | Parameter Description | Default Value |
|---|---|---|---|---|---|
print() |
Print the results to the terminal | format_json |
bool |
Whether to format the output content using JSON indentation |
True |
indent |
int |
Specify the indentation level to beautify the output JSON data, making it more readable, only effective when format_json is True |
4 | ||
ensure_ascii |
bool |
Control whether to escape non-ASCII characters to Unicode. If set to True, all non-ASCII characters will be escaped; False retains the original characters, only effective when format_json is True |
False
| ||
save_to_json() |
Save the results as a JSON file | save_path |
str |
The path to save the file. If it is a directory, the saved file name will be consistent with the input file name | None |
indent |
int |
Specify the indentation level to beautify the output JSON data, making it more readable, only effective when format_json is True |
4 | ||
ensure_ascii |
bool |
Control whether to escape non-ASCII characters to Unicode. If set to True, all non-ASCII characters will be escaped; False retains the original characters, only effective when format_json is True |
False
| ||
save_to_img() |
Save the results as an image file | save_path |
str |
The path to save the file. If it is a directory, the saved file name will be consistent with the input file name | None |
- Additionally, it supports obtaining the visualization image with results and the prediction results through attributes, as follows:
| Attribute | Attribute Description |
|---|---|
json |
Get the prediction result in json format |
img |
Get the visualization image in dict format |
For more information on using PaddleX's single-model inference APIs, please refer to the PaddleX Single-Model Python Script Usage Instructions.
IV. Custom Development¶
If you are seeking higher accuracy from existing models, you can use PaddleX's custom development capabilities to develop better instance segmentation models. Before using PaddleX to develop instance segmentation models, please ensure that you have installed the relevant model training plugins for segmentation in PaddleX. The installation process can be found in the custom development section of the PaddleX Local Installation Guide.
4.1 Data Preparation¶
Before model training, it is necessary to prepare the corresponding dataset for each task module. PaddleX provides data verification functionality for each module, and only data that passes the verification can be used for model training. Additionally, PaddleX provides demo datasets for each module, allowing you to complete subsequent development based on the officially provided demo data. If you wish to use a private dataset for subsequent model training, you can refer to the PaddleX Instance Segmentation Task Module Data Annotation Tutorial.
4.1.1 Download Demo Data¶
You can download the demo dataset to a specified folder using the following command:
wget https://paddle-model-ecology.bj.bcebos.com/paddlex/data/instance_seg_coco_examples.tar -P ./dataset
tar -xf ./dataset/instance_seg_coco_examples.tar -C ./dataset/
4.1.2 Data Verification¶
Data verification can be completed with a single command:
python main.py -c paddlex/configs/modules/instance_segmentation/Mask-RT-DETR-L.yaml \
-o Global.mode=check_dataset \
-o Global.dataset_dir=./dataset/instance_seg_coco_examples
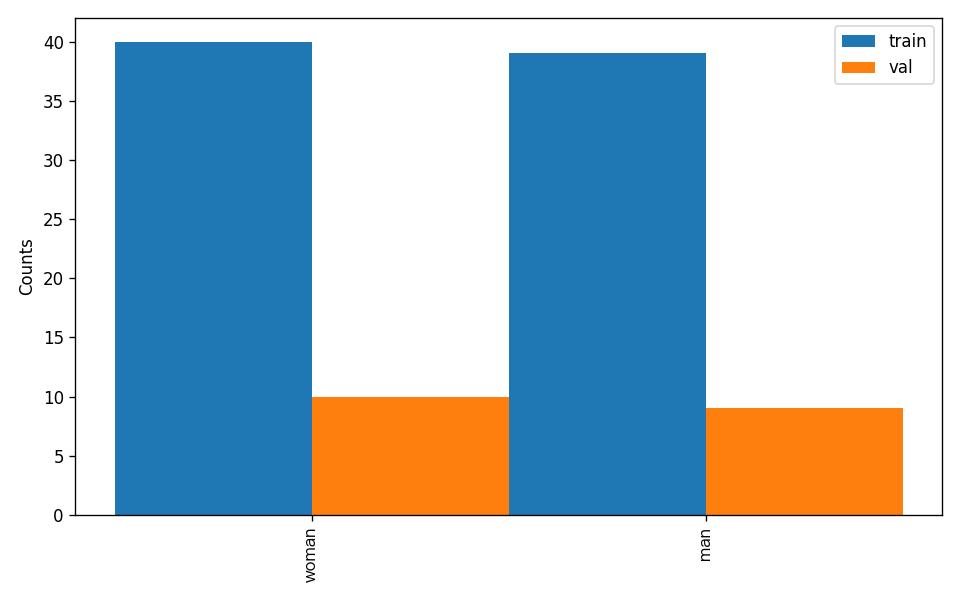
After executing the above command, PaddleX will validate the dataset and summarize its basic information. If the command runs successfully, it will print Check dataset passed ! in the log. The validation results file is saved in ./output/check_dataset_result.json, and related outputs are saved in the ./output/check_dataset directory in the current directory, including visual examples of sample images and sample distribution histograms.
👉 Details of Validation Results (Click to Expand)
The specific content of the validation result file is:
{
"done_flag": true,
"check_pass": true,
"attributes": {
"num_classes": 2,
"train_samples": 79,
"train_sample_paths": [
"check_dataset/demo_img/pexels-photo-634007.jpeg",
"check_dataset/demo_img/pexels-photo-59576.png"
],
"val_samples": 19,
"val_sample_paths": [
"check_dataset/demo_img/peasant-farmer-farmer-romania-botiza-47862.jpeg",
"check_dataset/demo_img/pexels-photo-715546.png"
]
},
"analysis": {
"histogram": "check_dataset/histogram.png"
},
"dataset_path": "instance_seg_coco_examples",
"show_type": "image",
"dataset_type": "COCOInstSegDataset"
}
In the above verification results, check_pass being True indicates that the dataset format meets the requirements. Explanations for other indicators are as follows:
attributes.num_classes: The number of classes in this dataset is 2;attributes.train_samples: The number of training samples in this dataset is 79;attributes.val_samples: The number of validation samples in this dataset is 19;attributes.train_sample_paths: A list of relative paths to the visualized training samples in this dataset;attributes.val_sample_paths: A list of relative paths to the visualized validation samples in this dataset; Additionally, the dataset verification also analyzes the distribution of sample numbers across all categories in the dataset and generates a distribution histogram (histogram.png):
4.1.3 Dataset Format Conversion/Dataset Splitting (Optional)¶
After completing data verification, you can convert the dataset format or re-split the training/validation ratio by modifying the configuration file or appending hyperparameters.
👉 Details of Format Conversion/Dataset Splitting (Click to Expand)
(1) Dataset Format Conversion
The instance segmentation task supports converting LabelMe format to COCO format. The parameters for dataset format conversion can be set by modifying the fields under CheckDataset in the configuration file. Below are some example explanations for some of the parameters in the configuration file:
CheckDataset:convert:enable: Whether to perform dataset format conversion. Set toTrueto enable dataset format conversion, default isFalse;src_dataset_type: If dataset format conversion is performed, the source dataset format needs to be set. The available source format isLabelMe; For example, if you want to convert aLabelMedataset toCOCOformat, you need to modify the configuration file as follows:
cd /path/to/paddlex
wget https://paddle-model-ecology.bj.bcebos.com/paddlex/data/instance_seg_labelme_examples.tar -P ./dataset
tar -xf ./dataset/instance_seg_labelme_examples.tar -C ./dataset/
......
CheckDataset:
......
convert:
enable: True
src_dataset_type: LabelMe
......
Then execute the command:
python main.py -c paddlex/configs/modules/instance_segmentation/Mask-RT-DETR-L.yaml\
-o Global.mode=check_dataset \
-o Global.dataset_dir=./dataset/instance_seg_labelme_examples
After the data conversion is executed, the original annotation files will be renamed to xxx.bak in the original path.
The above parameters also support being set by appending command line arguments:
python main.py -c paddlex/configs/modules/instance_segmentation/Mask-RT-DETR-L.yaml\
-o Global.mode=check_dataset \
-o Global.dataset_dir=./dataset/instance_seg_labelme_examples \
-o CheckDataset.convert.enable=True \
-o CheckDataset.convert.src_dataset_type=LabelMe
(2) Dataset Splitting
The parameters for dataset splitting can be set by modifying the fields under CheckDataset in the configuration file. Some example explanations for the parameters in the configuration file are as follows:
CheckDataset:split:enable: Whether to re-split the dataset. When set toTrue, the dataset will be re-split. The default isFalse;train_percent: If the dataset is to be re-split, the percentage of the training set needs to be set. The type is any integer between 0-100, and the sum withval_percentmust be 100;
For example, if you want to re-split the dataset with a 90% training set and a 10% validation set, you need to modify the configuration file as follows:
......
CheckDataset:
......
split:
enable: True
train_percent: 90
val_percent: 10
......
Then execute the command:
python main.py -c paddlex/configs/modules/instance_segmentation/Mask-RT-DETR-L.yaml \
-o Global.mode=check_dataset \
-o Global.dataset_dir=./dataset/instance_seg_labelme_examples
After data splitting, the original annotation files will be renamed as xxx.bak in the original path.
The above parameters can also be set by appending command line arguments:
python main.py -c paddlex/configs/modules/instance_segmentation/Mask-RT-DETR-L.yaml \
-o Global.mode=check_dataset \
-o Global.dataset_dir=./dataset/instance_seg_labelme_examples \
-o CheckDataset.split.enable=True \
-o CheckDataset.split.train_percent=90 \
-o CheckDataset.split.val_percent=10
4.2 Model Training¶
A single command can complete model training. Taking the training of the instance segmentation model Mask-RT-DETR-L as an example:
python main.py -c paddlex/configs/modules/instance_segmentation/Mask-RT-DETR-L.yaml \
-o Global.mode=train \
-o Global.dataset_dir=./dataset/instance_seg_coco_examples
- Specify the path to the
.yamlconfiguration file of the model (here it isMask-RT-DETR-L.yaml,When training other models, you need to specify the corresponding configuration files. The relationship between the model and configuration files can be found in the PaddleX Model List (CPU/GPU)) - Specify the mode as model training:
-o Global.mode=train - Specify the path to the training dataset:
-o Global.dataset_dir - Other related parameters can be set by modifying the fields under
GlobalandTrainin the.yamlconfiguration file, or adjusted by appending parameters in the command line. For example, to specify the first 2 GPUs for training:-o Global.device=gpu:0,1; to set the number of training epochs to 10:-o Train.epochs_iters=10. For more modifiable parameters and their detailed explanations, refer to the PaddleX Common Configuration File Parameters Instructions. - New Feature: Paddle 3.0 support CINN (Compiler Infrastructure for Neural Networks) to accelerate training speed when using GPU device. Please specify
-o Train.dy2st=Trueto enable it.
👉 More Details (Click to Expand)
- During model training, PaddleX automatically saves the model weight files, with the default being
output. If you need to specify a save path, you can set it through the-o Global.outputfield in the configuration file. - PaddleX shields you from the concepts of dynamic graph weights and static graph weights. During model training, both dynamic and static graph weights are produced, and static graph weights are selected by default for model inference.
-
After completing the model training, all outputs are saved in the specified output directory (default is
./output/), typically including: -
train_result.json: Training result record file, recording whether the training task was completed normally, as well as the output weight metrics, related file paths, etc.; train.log: Training log file, recording changes in model metrics and loss during training;config.yaml: Training configuration file, recording the hyperparameter configuration for this training session;.pdparams,.pdema,.pdopt.pdstate,.pdiparams,.json: Model weight-related files, including network parameters, optimizer, EMA, static graph network parameters, static graph network structure, etc.;- Notice: Since Paddle 3.0.0, the format of storing static graph network structure has changed to json(the current
.jsonfile) from protobuf(the former.pdmodelfile) to be compatible with PIR and more flexible and scalable.
4.3 Model Evaluation¶
After completing model training, you can evaluate the specified model weights file on the validation set to verify the model's accuracy. Using PaddleX for model evaluation can be done with a single command:
python main.py -c paddlex/configs/modules/instance_segmentation/Mask-RT-DETR-L.yaml \
-o Global.mode=evaluate \
-o Global.dataset_dir=./dataset/instance_seg_coco_examples
- Specify the
.yamlconfiguration file path for the model (here it isMask-RT-DETR-L.yaml) - Specify the mode as model evaluation:
-o Global.mode=evaluate - Specify the path to the validation dataset:
-o Global.dataset_dir. Other related parameters can be set by modifying theGlobalandEvaluatefields in the.yamlconfiguration file. For details, refer to PaddleX Common Model Configuration File Parameter Description.
👉 More Details (Click to Expand)
When evaluating the model, you need to specify the model weights file path. Each configuration file has a default weight save path built-in. If you need to change it, simply set it by appending a command line parameter, such as -o Evaluate.weight_path=./output/best_model/best_model.pdparams.
After completing the model evaluation, an evaluate_result.json file will be generated, which records the evaluation results, specifically whether the evaluation task was completed successfully and the model's evaluation metrics, including AP.
4.4 Model Inference and Model Integration¶
After completing model training and evaluation, you can use the trained model weights for inference prediction or Python integration.
4.4.1 Model Inference¶
To perform inference prediction via the command line, simply use the following command. Before running the following code, please download the demo image to your local machine.
python main.py -c paddlex/configs/modules/instance_segmentation/Mask-RT-DETR-L.yaml \
-o Global.mode=predict \
-o Predict.model_dir="./output/best_model/inference" \
-o Predict.input="general_instance_segmentation_004.png"
- Specify the
.yamlconfiguration file path of the model (here it'sMask-RT-DETR-L.yaml) - Specify the mode as model inference prediction:
-o Global.mode=predict - Specify the model weights path:
-o Predict.model_dir="./output/best_model/inference" - Specify the input data path:
-o Predict.input="...". Other related parameters can be set by modifying theGlobalandPredictfields in the.yamlconfiguration file. For details, refer to PaddleX Common Model Configuration File Parameter Description.
4.4.2 Model Integration¶
The model can be directly integrated into the PaddleX Pipeline or into your own project.
1.Pipeline Integration
The instance segmentation module can be integrated into the General Instance Segmentation Pipeline of PaddleX. Simply replace the model path to update the instance segmentation module of the relevant pipeline.
2.Module Integration The weights you produce can be directly integrated into the instance segmentation module. Refer to the Python example code in Quick Integration , and simply replace the model with the path to your trained model.
You can also use the PaddleX high-performance inference plugin to optimize the inference process of your model and further improve efficiency. For detailed procedures, please refer to the PaddleX High-Performance Inference Guide.