Semantic Segmentation Module Development Tutorial¶
I. Overview¶
Semantic segmentation is a technique in computer vision that classifies each pixel in an image, dividing the image into distinct semantic regions, with each region corresponding to a specific category. This technique generates detailed segmentation maps, clearly revealing objects and their boundaries in the image, providing powerful support for image analysis and understanding.
II. Supported Model List¶
| Model Name | Model Download Link | mIoU (%) | GPU Inference Time (ms) | CPU Inference Time (ms) | Model Size (M) |
|---|---|---|---|---|---|
| OCRNet_HRNet-W48 | Inference Model/Trained Model | 82.15 | 78.9976 | 2226.95 | 249.8 M |
| PP-LiteSeg-T | Inference Model/Trained Model | 73.10 | 7.6827 | 138.683 | 28.5 M |
❗ The above list features the 2 core models that the image classification module primarily supports. In total, this module supports 18 models. The complete list of models is as follows:
👉Model List Details
| Model Name | Model Download Link | mIoU (%) | GPU Inference Time (ms) | CPU Inference Time (ms) | Model Size (M) |
|---|---|---|---|---|---|
| Deeplabv3_Plus-R50 | Inference Model/Trained Model | 80.36 | 61.0531 | 1513.58 | 94.9 M |
| Deeplabv3_Plus-R101 | Inference Model/Trained Model | 81.10 | 100.026 | 2460.71 | 162.5 M |
| Deeplabv3-R50 | Inference Model/Trained Model | 79.90 | 82.2631 | 1735.83 | 138.3 M |
| Deeplabv3-R101 | Inference Model/Trained Model | 80.85 | 121.492 | 2685.51 | 205.9 M |
| OCRNet_HRNet-W18 | Inference Model/Trained Model | 80.67 | 48.2335 | 906.385 | 43.1 M |
| OCRNet_HRNet-W48 | Inference Model/Trained Model | 82.15 | 78.9976 | 2226.95 | 249.8 M |
| PP-LiteSeg-T | Inference Model/Trained Model | 73.10 | 7.6827 | 138.683 | 28.5 M |
| PP-LiteSeg-B | Inference Model/Trained Model | 75.25 | 10.9935 | 194.727 | 47.0 M |
| SegFormer-B0 (slice) | Inference Model/Trained Model | 76.73 | 11.1946 | 268.929 | 13.2 M |
| SegFormer-B1 (slice) | Inference Model/Trained Model | 78.35 | 17.9998 | 403.393 | 48.5 M |
| SegFormer-B2 (slice) | Inference Model/Trained Model | 81.60 | 48.0371 | 1248.52 | 96.9 M |
| SegFormer-B3 (slice) | Inference Model/Trained Model | 82.47 | 64.341 | 1666.35 | 167.3 M |
| SegFormer-B4 (slice) | Inference Model/Trained Model | 82.38 | 82.4336 | 1995.42 | 226.7 M |
| SegFormer-B5 (slice) | Inference Model/Trained Model | 82.58 | 97.3717 | 2420.19 | 229.7 M |
The accuracy metrics of the above models are measured on the Cityscapes dataset. GPU inference time is based on an NVIDIA Tesla T4 machine with FP32 precision. CPU inference speed is based on an Intel(R) Xeon(R) Gold 5117 CPU @ 2.00GHz with 8 threads and FP32 precision.
| Model Name | Model Download Link | mIoU (%) | GPU Inference Time (ms) | CPU Inference Time | Model Size (M) |
|---|---|---|---|---|---|
| SeaFormer_base(slice) | Inference Model/Trained Model | 40.92 | 24.4073 | 397.574 | 30.8 M |
| SeaFormer_large (slice) | Inference Model/Trained Model | 43.66 | 27.8123 | 550.464 | 49.8 M |
| SeaFormer_small (slice) | Inference Model/Trained Model | 38.73 | 19.2295 | 358.343 | 14.3 M |
| SeaFormer_tiny (slice) | Inference Model/Trained Model | 34.58 | 13.9496 | 330.132 | 6.1M |
The accuracy metrics of the SeaFormer series models are measured on the ADE20k dataset. GPU inference time is based on an NVIDIA Tesla T4 machine with FP32 precision. CPU inference speed is based on an Intel(R) Xeon(R) Gold 5117 CPU @ 2.00GHz with 8 threads and FP32 precision.
III. Quick Integration¶
❗ Before quick integration, please install the PaddleX wheel package. For detailed instructions, refer to the PaddleX Local Installation Guide
Just a few lines of code can complete the inference of the Semantic Segmentation module, allowing you to easily switch between models under this module. You can also integrate the model inference of the the Semantic Segmentation module into your project. Before running the following code, please download the demo image to your local machine.
from paddlex import create_model
model = create_model("PP-LiteSeg-T")
output = model.predict("general_semantic_segmentation_002.png", batch_size=1)
for res in output:
res.print(json_format=False)
res.save_to_img("./output/")
res.save_to_json("./output/res.json")
IV. Custom Development¶
If you seek higher accuracy, you can leverage PaddleX's custom development capabilities to develop better Semantic Segmentation models. Before developing a Semantic Segmentation model with PaddleX, ensure you have installed PaddleClas plugin for PaddleX. The installation process can be found in the custom development section of the PaddleX Local Installation Tutorial.
4.1 Dataset Preparation¶
Before model training, you need to prepare a dataset for the task. PaddleX provides data validation functionality for each module. Only data that passes validation can be used for model training. Additionally, PaddleX provides demo datasets for each module, which you can use to complete subsequent development. If you wish to use private datasets for model training, refer to PaddleX Semantic Segmentation Task Module Data Preparation Tutorial.
4.1.1 Demo Data Download¶
You can download the demo dataset to a specified folder using the following commands:
wget https://paddle-model-ecology.bj.bcebos.com/paddlex/data/seg_optic_examples.tar -P ./dataset
tar -xf ./dataset/seg_optic_examples.tar -C ./dataset/
4.1.2 Data Validation¶
Data validation can be completed with a single command:
python main.py -c paddlex/configs/semantic_segmentation/PP-LiteSeg-T.yaml \
-o Global.mode=check_dataset \
-o Global.dataset_dir=./dataset/seg_optic_examples
After executing the above command, PaddleX will verify the dataset and collect basic information about it. Once the command runs successfully, a message saying Check dataset passed ! will be printed in the log. The verification results will be saved in ./output/check_dataset_result.json, and related outputs will be stored in the ./output/check_dataset directory, including visual examples of sample images and a histogram of sample distribution.
👉 Verification Result Details (click to expand)
The specific content of the verification result file is:
{
"done_flag": true,
"check_pass": true,
"attributes": {
"train_sample_paths": [
"check_dataset/demo_img/P0005.jpg",
"check_dataset/demo_img/P0050.jpg"
],
"train_samples": 267,
"val_sample_paths": [
"check_dataset/demo_img/N0139.jpg",
"check_dataset/demo_img/P0137.jpg"
],
"val_samples": 76,
"num_classes": 2
},
"analysis": {
"histogram": "check_dataset/histogram.png"
},
"dataset_path": "./dataset/seg_optic_examples",
"show_type": "image",
"dataset_type": "SegDataset"
}
The verification results above indicate that check_pass being True means the dataset format meets the requirements. Explanations for other indicators are as follows:
attributes.num_classes: The number of classes in this dataset is 2;attributes.train_samples: The number of training samples in this dataset is 267;attributes.val_samples: The number of validation samples in this dataset is 76;attributes.train_sample_paths: A list of relative paths to the visualization images of training samples in this dataset;attributes.val_sample_paths: A list of relative paths to the visualization images of validation samples in this dataset;
The dataset verification also analyzes the distribution of sample numbers across all classes and plots a histogram (histogram.png):
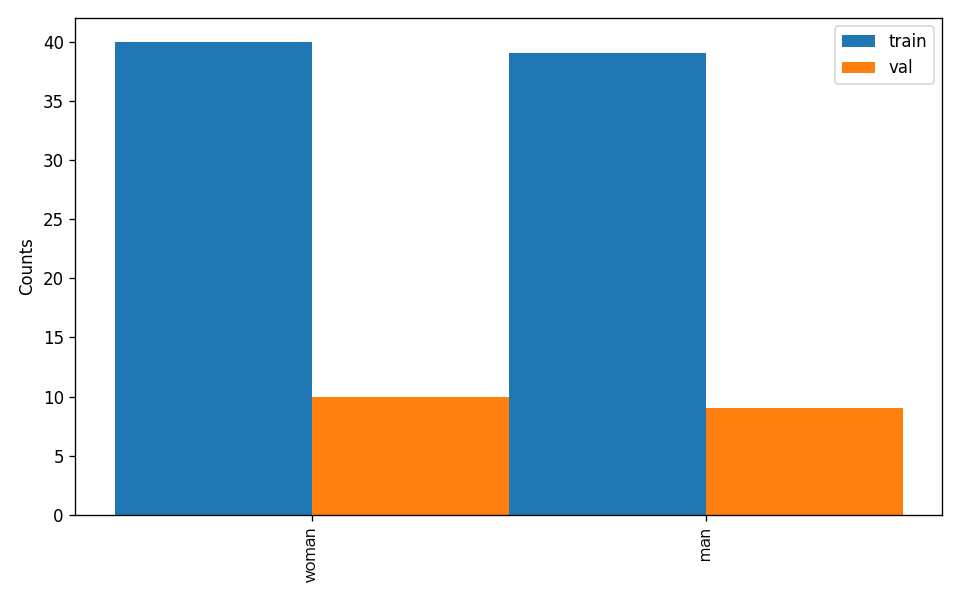
4.1.3 Dataset Format Conversion/Dataset Splitting (Optional) (Click to Expand)¶
👉 Details on Format Conversion/Dataset Splitting (Click to Expand)
After completing dataset verification, you can convert the dataset format or re-split the training/validation ratio by modifying the configuration file or appending hyperparameters.
(1) Dataset Format Conversion
Semantic segmentation supports converting LabelMe format datasets to the required format.
Parameters related to dataset verification can be set by modifying the CheckDataset fields in the configuration file. Example explanations for some parameters in the configuration file are as follows:
CheckDataset:convert:enable: Whether to enable dataset format conversion, supportingLabelMeformat conversion, default isFalse;src_dataset_type: If dataset format conversion is enabled, the source dataset format needs to be set, default isnull, and the supported source dataset format isLabelMe;
For example, if you want to convert a LabelMe format dataset, you can download a sample LabelMe format dataset as follows:
wget https://paddle-model-ecology.bj.bcebos.com/paddlex/data/seg_dataset_to_convert.tar -P ./dataset
tar -xf ./dataset/seg_dataset_to_convert.tar -C ./dataset/
After downloading, modify the paddlex/configs/semantic_segmentation/PP-LiteSeg-T.yaml configuration as follows:
......
CheckDataset:
......
convert:
enable: True
src_dataset_type: LabelMe
......
Then execute the command:
python main.py -c paddlex/configs/semantic_segmentation/PP-LiteSeg-T.yaml \
-o Global.mode=check_dataset \
-o Global.dataset_dir=./dataset/seg_dataset_to_convert
Of course, the above parameters also support being set by appending command-line arguments. For a LabelMe format dataset, the command is:
python main.py -c paddlex/configs/semantic_segmentation/PP-LiteSeg-T.yaml \
-o Global.mode=check_dataset \
-o Global.dataset_dir=./dataset/seg_dataset_to_convert \
-o CheckDataset.convert.enable=True \
-o CheckDataset.convert.src_dataset_type=LabelMe
(2) Dataset Splitting
Parameters for dataset splitting can be set by modifying the CheckDataset fields in the configuration file. Example explanations for some parameters in the configuration file are as follows:
CheckDataset:split:enable: Whether to enable re-splitting the dataset, set toTrueto perform dataset splitting, default isFalse;train_percent: If re-splitting the dataset, set the percentage of the training set, which should be an integer between 0 and 100, ensuring the sum withval_percentis 100;
For example, if you want to re-split the dataset with a 90% training set and a 10% validation set, modify the configuration file as follows:
......
CheckDataset:
......
split:
enable: True
train_percent: 90
val_percent: 10
......
Then execute the command:
python main.py -c paddlex/configs/semantic_segmentation/PP-LiteSeg-T.yaml \
-o Global.mode=check_dataset \
-o Global.dataset_dir=./dataset/seg_optic_examples
After dataset splitting, the original annotation files will be renamed to xxx.bak in the original path.
The above parameters also support setting through appending command line arguments:
python main.py -c paddlex/configs/semantic_segmentation/PP-LiteSeg-T.yaml \
-o Global.mode=check_dataset \
-o Global.dataset_dir=./dataset/seg_optic_examples \
-o CheckDataset.split.enable=True \
-o CheckDataset.split.train_percent=90 \
-o CheckDataset.split.val_percent=10
4.2 Model Training¶
Model training can be completed with just one command. Here, we use the semantic segmentation model (PP-LiteSeg-T) as an example:
python main.py -c paddlex/configs/semantic_segmentation/PP-LiteSeg-T.yaml \
-o Global.mode=train \
-o Global.dataset_dir=./dataset/seg_optic_examples
You need to follow these steps:
- Specify the
.yamlconfiguration file path for the model (here it'sPP-LiteSeg-T.yaml,When training other models, you need to specify the corresponding configuration files. The relationship between the model and configuration files can be found in the PaddleX Model List (CPU/GPU)). - Set the mode to model training:
-o Global.mode=train - Specify the training dataset path:
-o Global.dataset_dir
Other related parameters can be set by modifying the Global and Train fields in the .yaml configuration file, or adjusted by appending parameters in the command line. For example, to train using the first two GPUs: -o Global.device=gpu:0,1; to set the number of training epochs to 10: -o Train.epochs_iters=10. For more modifiable parameters and their detailed explanations, refer to the PaddleX Common Configuration Parameters Documentation.
👉 More Details (Click to Expand)
- During model training, PaddleX automatically saves model weight files, with the default path being
output. To specify a different save path, use the-o Global.outputfield in the configuration file. - PaddleX abstracts the concepts of dynamic graph weights and static graph weights from you. During model training, both dynamic and static graph weights are produced, and static graph weights are used by default for model inference.
-
After model training, all outputs are saved in the specified output directory (default is
./output/), typically including: -
train_result.json: Training result record file, including whether the training task completed successfully, produced weight metrics, and related file paths. train.log: Training log file, recording model metric changes, loss changes, etc.config.yaml: Training configuration file, recording the hyperparameters used for this training session..pdparams,.pdema,.pdopt.pdstate,.pdiparams,.pdmodel: Model weight-related files, including network parameters, optimizer, EMA, static graph network parameters, and static graph network structure.
4.3 Model Evaluation¶
After model training, you can evaluate the specified model weights on the validation set to verify model accuracy. Using PaddleX for model evaluation requires just one command:
python main.py -c paddlex/configs/semantic_segmentation/PP-LiteSeg-T.yaml \
-o Global.mode=evaluate \
-o Global.dataset_dir=./dataset/seg_optic_examples
Similar to model training, follow these steps:
- Specify the
.yamlconfiguration file path for the model (here it'sPP-LiteSeg-T.yaml). - Set the mode to model evaluation:
-o Global.mode=evaluate - Specify the validation dataset path:
-o Global.dataset_dir
Other related parameters can be set by modifying the Global and Evaluate fields in the .yaml configuration file. For more details, refer to the PaddleX Common Configuration Parameters Documentation.
👉 More Details (Click to Expand)
When evaluating the model, you need to specify the model weight file path. Each configuration file has a default weight save path. If you need to change it, simply append the command line parameter, e.g., -o Evaluate.weight_path=./output/best_model/best_model.pdparams.
After model evaluation, the following outputs are typically produced:
evaluate_result.json: Records the evaluation results, specifically whether the evaluation task completed successfully and the model's evaluation metrics, including mIoU.
4.4 Model Inference and Integration¶
After model training and evaluation, you can use the trained model weights for inference predictions or Python integration.
4.4.1 Model Inference¶
To perform inference predictions via the command line, use the following command. Before running the following code, please download the demo image to your local machine.
python main.py -c paddlex/configs/semantic_segmentation/PP-LiteSeg-T.yaml \
-o Global.mode=predict \
-o Predict.model_dir="./output/best_model" \
-o Predict.input="general_semantic_segmentation_002.png"
Similar to model training and evaluation, the following steps are required:
-
Specify the
.yamlconfiguration file path of the model (here it'sPP-LCNet_x1_0_doc_ori.yaml) -
Set the mode to model inference prediction:
-o Global.mode=predict -
Specify the model weights path: -o Predict.model_dir="./output/best_accuracy/inference"
Specify the input data path: -o Predict.inputh="..." Other related parameters can be set by modifying the fields under Global and Predict in the .yaml configuration file. For details, refer to PaddleX Common Model Configuration File Parameter Description.
Alternatively, you can use the PaddleX wheel package for inference, easily integrating the model into your own projects.
4.4.2 Model Integration¶
The model can be directly integrated into the PaddleX pipeline or into your own projects.
- Pipeline Integration
The document semantic segmentation module can be integrated into PaddleX pipelines such as the Semantic Segmentation Pipeline (Seg). Simply replace the model path to update the The document semantic segmentation module's model.
- Module Integration
The weights you produce can be directly integrated into the semantic segmentation module. You can refer to the Python sample code in Quick Integration and just replace the model with the path to the model you trained.