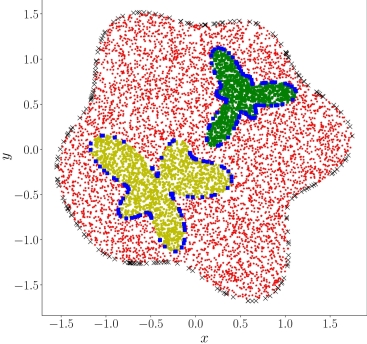
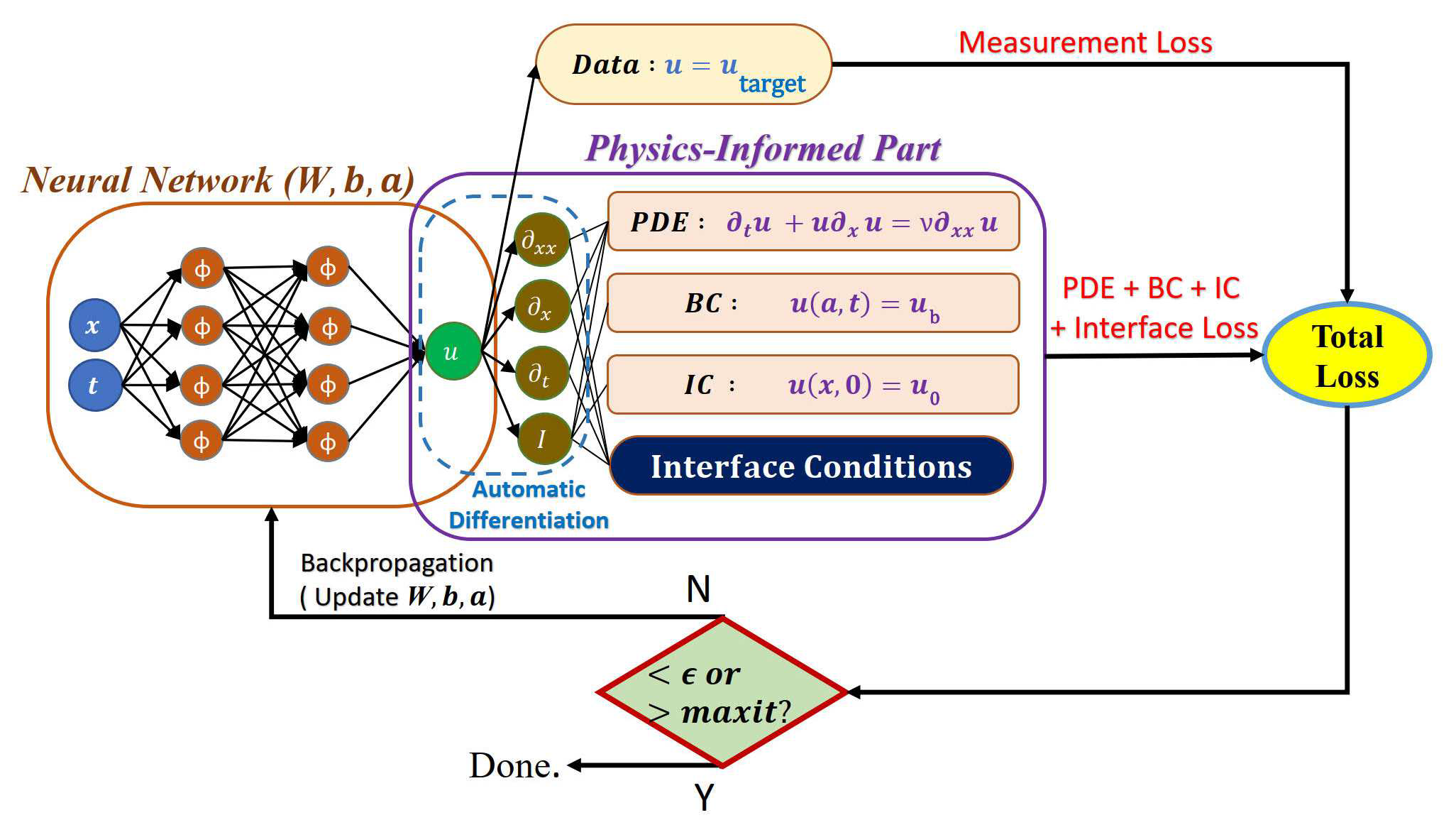
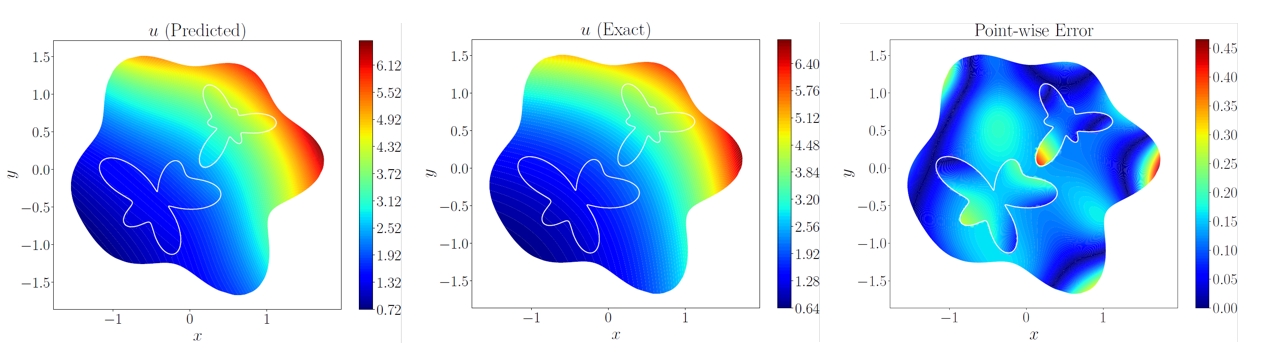
# Copyright (c) 2024 PaddlePaddle Authors. All Rights Reserved.
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
# http://www.apache.org/licenses/LICENSE-2.0
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
from typing import Callable
from typing import Dict
from typing import List
from typing import Tuple
import hydra
import model
import numpy as np
import paddle
import plotting
from omegaconf import DictConfig
import ppsci
# For the use of the second derivative: paddle.cos
paddle.framework.core.set_prim_eager_enabled(True)
def _xpinn_loss(
training_pres: List[List[paddle.Tensor]] = None,
training_exacts: List[paddle.Tensor] = None,
training_weight: float = 1,
residual_inputs: List[List[paddle.Tensor]] = None,
residual_pres: List[paddle.Tensor] = None,
residual_weight: float = 1,
interface_inputs: List[List[paddle.Tensor]] = None,
interface_pres: List[paddle.Tensor] = None,
interface_weight: float = 1,
interface_neigh_pres: List[List[paddle.Tensor]] = None,
interface_neigh_weight: float = 1,
residual_func: Callable = lambda x, y: x - y,
) -> float:
"""XPINNs loss function for subdomain
`loss = W_u_q * MSE_u_q + W_F_q * MSE_F_q + W_I_q * MSE_avg_q + W_I_F_q * MSE_R`
`W_u_q * MSE_u_q` is data mismatch item.
`W_F_q * MSE_F_q` is residual item.
`W_I_q * MSE_avg_q` is interface item.
`W_I_F_q * MSE_R` is interface residual item.
Args:
training_pres (List[List[paddle.Tensor]], optional): the prediction result for training points input. Defaults to None.
training_exacts (List[paddle.Tensor], optional): the exact result for training points input. Defaults to None.
training_weight (float, optional): the weight of data mismatch item. Defaults to 1.
residual_inputs (List[List[paddle.Tensor]], optional): residual points input. Defaults to None.
residual_pres (List[paddle.Tensor], optional): the prediction result for residual points input. Defaults to None.
residual_weight (float, optional): the weight of residual item. Defaults to 1.
interface_inputs (List[List[paddle.Tensor]], optional): the prediction result for interface points input. Defaults to None.
interface_pres (List[paddle.Tensor], optional): the prediction result for interface points input. Defaults to None.
interface_weight (float, optional): the weight of iinterface item. Defaults to 1.
interface_neigh_pres (List[List[paddle.Tensor]], optional): the prediction result of neighbouring subdomain model for interface points input. Defaults to None.
interface_neigh_weight (float, optional): the weight of interface residual term. Defaults to 1.
residual_func (Callable, optional): residual calculation function. Defaults to lambda x,y : x - y.
"""
def _get_grad(outputs: paddle.Tensor, inputs: paddle.Tensor) -> paddle.Tensor:
grad = paddle.grad(outputs, inputs, retain_graph=True, create_graph=True)
return grad[0]
def _get_second_derivatives(
outputs_list: List[paddle.Tensor],
inputs_list: List[List[paddle.Tensor]],
) -> Tuple[List[List[paddle.Tensor]], List[List[paddle.Tensor]]]:
d1_list = [
[_get_grad(_out, _in) for _in in _ins]
for _out, _ins in zip(outputs_list, inputs_list)
]
d2_list = [
[_get_grad(_d1, _in) for _d1, _in in zip(d1s_, _ins)]
for d1s_, _ins in zip(d1_list, inputs_list)
]
return d2_list
residual_u_d2_list = _get_second_derivatives(residual_pres, residual_inputs)
interface_u_d2_list = _get_second_derivatives(interface_pres, interface_inputs)
interface_neigh_u_d2_list = _get_second_derivatives(
interface_neigh_pres, interface_inputs
)
MSE_u_q = 0
if training_pres is not None:
for _pre, _exact in zip(training_pres, training_exacts):
MSE_u_q += training_weight * paddle.mean(paddle.square(_pre - _exact))
MSE_F_q = 0
if residual_inputs is not None:
for _ins, _d2 in zip(residual_inputs, residual_u_d2_list):
MSE_F_q += residual_weight * paddle.mean(
paddle.square(residual_func(_d2, _ins))
)
MSE_avg_q = 0
MSE_R = 0
if interface_inputs is not None:
for _ins, _pre, _n_pres in zip(
interface_inputs, interface_pres, interface_neigh_pres
):
pre_list = [_pre] + _n_pres
pre_avg = paddle.add_n(pre_list) / len(pre_list)
MSE_avg_q += interface_weight * paddle.mean(paddle.square(_pre - pre_avg))
for _ins, _d2, _n_d2 in zip(
interface_inputs, interface_u_d2_list, interface_neigh_u_d2_list
):
MSE_R += interface_neigh_weight * paddle.mean(
paddle.square(residual_func(_d2, _ins) - residual_func(_n_d2, _ins))
)
return MSE_u_q + MSE_F_q + MSE_avg_q + MSE_R
def loss_fun(
output_dict: Dict[str, paddle.Tensor],
label_dict: Dict[str, paddle.Tensor],
*args,
) -> float:
def residual_func(output_der: paddle.Tensor, input: paddle.Tensor) -> paddle.Tensor:
return paddle.add_n(output_der) - paddle.add_n(
[paddle.exp(_in) for _in in input]
)
# subdomain 1
loss1 = _xpinn_loss(
training_pres=[output_dict["boundary_u"]],
training_exacts=[label_dict["boundary_u_exact"]],
training_weight=20,
residual_inputs=[[output_dict["residual1_x"], output_dict["residual1_y"]]],
residual_pres=[output_dict["residual1_u"]],
residual_weight=1,
interface_inputs=[
[output_dict["interface1_x"], output_dict["interface1_y"]],
[output_dict["interface2_x"], output_dict["interface2_y"]],
],
interface_pres=[
output_dict["interface1_u_sub1"],
output_dict["interface2_u_sub1"],
],
interface_weight=20,
interface_neigh_pres=[
[output_dict["interface1_u_sub2"]],
[output_dict["interface2_u_sub3"]],
],
interface_neigh_weight=1,
residual_func=residual_func,
)
# subdomain 2
loss2 = _xpinn_loss(
residual_inputs=[[output_dict["residual2_x"], output_dict["residual2_y"]]],
residual_pres=[output_dict["residual2_u"]],
residual_weight=1,
interface_inputs=[[output_dict["interface1_x"], output_dict["interface1_y"]]],
interface_pres=[output_dict["interface1_u_sub1"]],
interface_weight=20,
interface_neigh_pres=[[output_dict["interface1_u_sub2"]]],
interface_neigh_weight=1,
residual_func=residual_func,
)
# subdomain 3
loss3 = _xpinn_loss(
residual_inputs=[[output_dict["residual3_x"], output_dict["residual3_y"]]],
residual_pres=[output_dict["residual3_u"]],
residual_weight=1,
interface_inputs=[[output_dict["interface2_x"], output_dict["interface2_y"]]],
interface_pres=[output_dict["interface2_u_sub1"]],
interface_weight=20,
interface_neigh_pres=[[output_dict["interface2_u_sub3"]]],
interface_neigh_weight=1,
residual_func=residual_func,
)
return {"residuals": loss1 + loss2 + loss3}
def eval_l2_rel_func(
output_dict: Dict[str, paddle.Tensor],
label_dict: Dict[str, paddle.Tensor],
*args,
) -> Dict[str, paddle.Tensor]:
u_pred = paddle.concat(
[
output_dict["residual1_u"],
output_dict["residual2_u"],
output_dict["residual3_u"],
]
)
# the shape of label_dict["residual_u_exact"] is [22387, 1], and be cut into [18211, 1] `_eval_by_dataset`(ppsci/solver/eval.py).
u_exact = paddle.concat(
[
label_dict["residual_u_exact"],
label_dict["residual2_u_exact"],
label_dict["residual3_u_exact"],
]
)
error_total = paddle.linalg.norm(
u_exact.flatten() - u_pred.flatten(), 2
) / paddle.linalg.norm(u_exact.flatten(), 2)
return {"l2_error": error_total}
def train(cfg: DictConfig):
# set training dataset transformation
def train_dataset_transform_func(
_input: Dict[str, np.ndarray],
_label: Dict[str, np.ndarray],
weight_: Dict[str, np.ndarray],
) -> Dict[str, np.ndarray]:
# Randomly select the residual points from sub-domains
id_x1 = np.random.choice(
_input["residual1_x"].shape[0],
cfg.MODEL.num_residual1_points,
replace=False,
)
_input["residual1_x"] = _input["residual1_x"][id_x1, :]
_input["residual1_y"] = _input["residual1_y"][id_x1, :]
id_x2 = np.random.choice(
_input["residual2_x"].shape[0],
cfg.MODEL.num_residual2_points,
replace=False,
)
_input["residual2_x"] = _input["residual2_x"][id_x2, :]
_input["residual2_y"] = _input["residual2_y"][id_x2, :]
id_x3 = np.random.choice(
_input["residual3_x"].shape[0],
cfg.MODEL.num_residual3_points,
replace=False,
)
_input["residual3_x"] = _input["residual3_x"][id_x3, :]
_input["residual3_y"] = _input["residual3_y"][id_x3, :]
# Randomly select boundary points
id_x4 = np.random.choice(
_input["boundary_x"].shape[0], cfg.MODEL.num_boundary_points, replace=False
)
_input["boundary_x"] = _input["boundary_x"][id_x4, :]
_input["boundary_y"] = _input["boundary_y"][id_x4, :]
_label["boundary_u_exact"] = _label["boundary_u_exact"][id_x4, :]
# Randomly select the interface points along two interfaces
id_xi1 = np.random.choice(
_input["interface1_x"].shape[0], cfg.MODEL.num_interface1, replace=False
)
_input["interface1_x"] = _input["interface1_x"][id_xi1, :]
_input["interface1_y"] = _input["interface1_y"][id_xi1, :]
id_xi2 = np.random.choice(
_input["interface2_x"].shape[0], cfg.MODEL.num_interface2, replace=False
)
_input["interface2_x"] = _input["interface2_x"][id_xi2, :]
_input["interface2_y"] = _input["interface2_y"][id_xi2, :]
return _input, _label, weight_
# set dataloader config
train_dataloader_cfg = {
"dataset": {
"name": "IterableMatDataset",
"file_path": cfg.DATA_FILE,
"input_keys": cfg.TRAIN.input_keys,
"label_keys": cfg.TRAIN.label_keys,
"alias_dict": cfg.TRAIN.alias_dict,
"transforms": (
{
"FunctionalTransform": {
"transform_func": train_dataset_transform_func,
},
},
),
}
}
layer_list = (
cfg.MODEL.layers1,
cfg.MODEL.layers2,
cfg.MODEL.layers3,
)
# set model
custom_model = model.Model(layer_list)
# set constraint
sup_constraint = ppsci.constraint.SupervisedConstraint(
train_dataloader_cfg,
ppsci.loss.FunctionalLoss(loss_fun),
{"residual1_u": lambda out: out["residual1_u"]},
name="sup_constraint",
)
constraint = {sup_constraint.name: sup_constraint}
# set validator
eval_dataloader_cfg = {
"dataset": {
"name": "IterableMatDataset",
"file_path": cfg.DATA_FILE,
"input_keys": cfg.TRAIN.input_keys,
"label_keys": cfg.EVAL.label_keys,
"alias_dict": cfg.EVAL.alias_dict,
}
}
sup_validator = ppsci.validate.SupervisedValidator(
eval_dataloader_cfg,
loss=ppsci.loss.FunctionalLoss(loss_fun),
output_expr={
"residual1_u": lambda out: out["residual1_u"],
"residual2_u": lambda out: out["residual2_u"],
"residual3_u": lambda out: out["residual3_u"],
},
metric={"L2Rel": ppsci.metric.FunctionalMetric(eval_l2_rel_func)},
name="sup_validator",
)
validator = {sup_validator.name: sup_validator}
# set optimizer
optimizer = ppsci.optimizer.Adam(cfg.TRAIN.learning_rate)(custom_model)
# initialize solver
solver = ppsci.solver.Solver(
custom_model,
constraint,
optimizer=optimizer,
validator=validator,
cfg=cfg,
)
solver.train()
solver.eval()
# visualize prediction
with solver.no_grad_context_manager(True):
for index, (_input, _label, _) in enumerate(sup_validator.data_loader):
u_exact = _label["residual_u_exact"]
output_ = custom_model(_input)
u_pred = paddle.concat(
[output_["residual1_u"], output_["residual2_u"], output_["residual3_u"]]
)
plotting.log_image(
residual1_x=_input["residual1_x"],
residual1_y=_input["residual1_y"],
residual2_x=_input["residual2_x"],
residual2_y=_input["residual2_y"],
residual3_x=_input["residual3_x"],
residual3_y=_input["residual3_y"],
interface1_x=_input["interface1_x"],
interface1_y=_input["interface1_y"],
interface2_x=_input["interface2_x"],
interface2_y=_input["interface2_y"],
boundary_x=_input["boundary_x"],
boundary_y=_input["boundary_y"],
residual_u_pred=u_pred,
residual_u_exact=u_exact,
)
def evaluate(cfg: DictConfig):
layer_list = (
cfg.MODEL.layers1,
cfg.MODEL.layers2,
cfg.MODEL.layers3,
)
custom_model = model.Model(layer_list)
# set validator
eval_dataloader_cfg = {
"dataset": {
"name": "IterableMatDataset",
"file_path": cfg.DATA_FILE,
"input_keys": cfg.TRAIN.input_keys,
"label_keys": cfg.EVAL.label_keys,
"alias_dict": cfg.EVAL.alias_dict,
}
}
sup_validator = ppsci.validate.SupervisedValidator(
eval_dataloader_cfg,
loss=ppsci.loss.FunctionalLoss(loss_fun),
output_expr={
"residual1_u": lambda out: out["residual1_u"],
"residual2_u": lambda out: out["residual2_u"],
"residual3_u": lambda out: out["residual3_u"],
},
metric={"L2Rel": ppsci.metric.FunctionalMetric(eval_l2_rel_func)},
name="sup_validator",
)
validator = {sup_validator.name: sup_validator}
# initialize solver
solver = ppsci.solver.Solver(
custom_model,
validator=validator,
cfg=cfg,
)
solver.eval()
# visualize prediction
with solver.no_grad_context_manager(True):
for index, (_input, _label, _) in enumerate(sup_validator.data_loader):
u_exact = _label["residual_u_exact"]
_output = custom_model(_input)
u_pred = paddle.concat(
[_output["residual1_u"], _output["residual2_u"], _output["residual3_u"]]
)
plotting.log_image(
residual1_x=_input["residual1_x"],
residual1_y=_input["residual1_y"],
residual2_x=_input["residual2_x"],
residual2_y=_input["residual2_y"],
residual3_x=_input["residual3_x"],
residual3_y=_input["residual3_y"],
interface1_x=_input["interface1_x"],
interface1_y=_input["interface1_y"],
interface2_x=_input["interface2_x"],
interface2_y=_input["interface2_y"],
boundary_x=_input["boundary_x"],
boundary_y=_input["boundary_y"],
residual_u_pred=u_pred,
residual_u_exact=u_exact,
)
@hydra.main(version_base=None, config_path="./conf", config_name="xpinn.yaml")
def main(cfg: DictConfig):
if cfg.mode == "train":
train(cfg)
elif cfg.mode == "eval":
evaluate(cfg)
else:
raise ValueError(f"cfg.mode should in ['train', 'eval'], but got '{cfg.mode}'")
if __name__ == "__main__":
main()